Table of Contents
AI-based Drug recommendation systems are rapidly becoming more important in the field of healthcare. With vast amounts of medical data and research available, it can be challenging for healthcare professionals to stay up-to-date with the latest treatments and medications.
AI-powered drug recommendation systems can provide valuable assistance to healthcare professionals in making more informed decisions regarding the best course of treatment for their patients.
One of the key benefits of drug recommendation using AI is that it can help identify potential drug interactions and adverse reactions. Such systems analyze patient data, including medical history and current medications, in order to identify potential issues and recommend appropriate alternatives.
Another benefit of AI-powered drug recommendation systems is that they can help reduce the risk of medication errors.
By automating the process of medication selection, dosing, and administration, these systems can ensure that patients receive the right medication in the right dose at the right time. This can result in better patient outcomes and a decrease in healthcare costs.
In this article, we will build a drug recommendation system using NLP and machine learning algorithms that will not only predict medical conditions but also recommend the top 3 drugs based on predicted medical conditions and top reviews and useful reviews count.
Overall Flow of the project
Let’s try to understand the overall flow of the drug recommendation system.
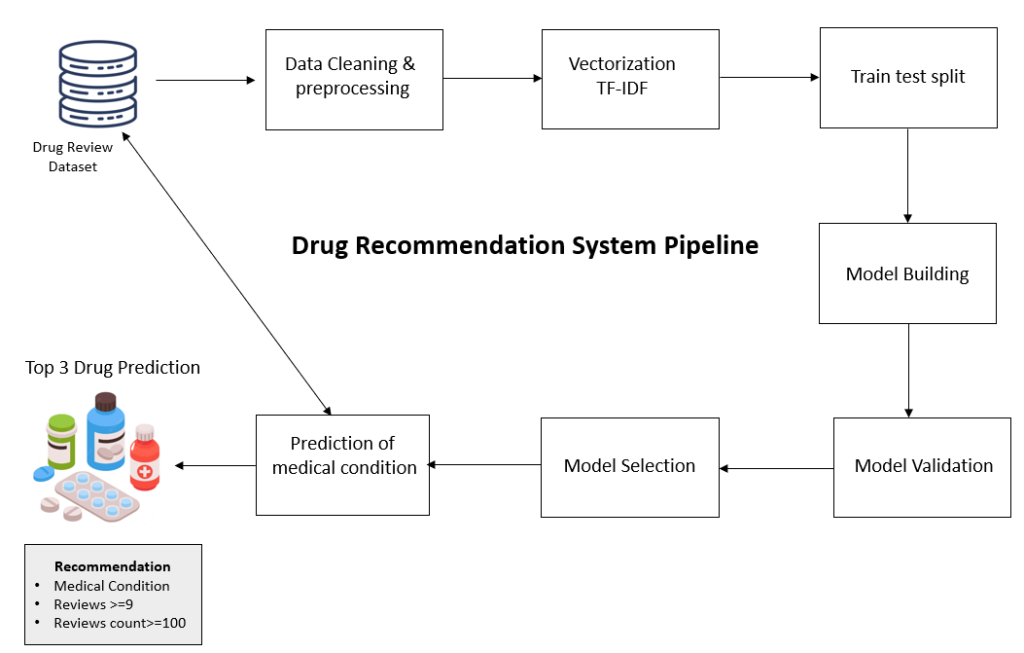
As per the flow of the project, we will first read the Drug Reviews dataset available on the UCI ML repository, then we will do text preprocessing and data cleaning to remove stop words, and special symbols, and then vectorize it to convert it into numerical format using TF-IDF. After vectorization, we will split the data into training and test set. Once splitting is done we will proceed to build a machine learning model and evaluate their performance. Next based on the performance criteria we will select next machine learning model for classifying medical conditions based on textual reviews of the users.
Based on the prediction of medical condition we will select the top 3 drugs from the dataset having ratings >=9 and total ratings >=100.
So, next start the process of building machine learning model for classifying medical conditions based on textual reviews.
Importing Libraries
import pandas as pd # data preprocessing
import itertools # confusion matrix
import string
import numpy as np
import seaborn as sns
from sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer
from sklearn.model_selection import train_test_split
from sklearn.linear_model import PassiveAggressiveClassifier
from sklearn.naive_bayes import MultinomialNB
from sklearn import metrics
import matplotlib.pyplot as plt
%matplotlib inline
# To show all the rows of pandas dataframe
pd.set_option('display.max_rows', None)Reading Dataset
In this project, we will be using the DrugReviews dataset available at UCI ML Repository.
The dataset consists of 1,61,297 reviews of the users with 7 columns. the snapshot of the dataset is shared below.

The dataset consists of drug names, medical conditions, textual reviews of the user, rating given by the user, date of the review, and a total number of useful count which means a total number of users who find the review useful.
Now, let’s check the distribution of medical conditions in the dataset.
df.condition.value_counts()
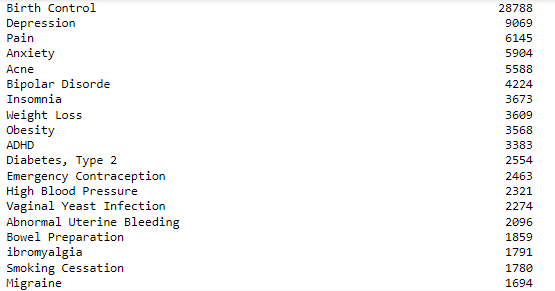
To make it simple we will select the top 4 medical conditions i.e., Birth Control, Depression, Diabetes, Type 2, and High Blood Pressure.
df_train = df[(df['condition']=='Birth Control') | (df['condition']=='Depression') | (df['condition']=='High Blood Pressure')|(df['condition']=='Diabetes, Type 2')]
After selecting the top 3 medical conditions, we will remove other features from the dataset. As for building a classifier, we need textual review and the medical condition as the target variable.
X = df_train.drop(['Unnamed: 0','drugName','rating','date','usefulCount'],axis=1)
Now, let’s check the distribution of medical conditions in dataset X.
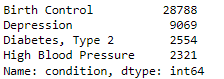
Now, let’s see the random sample reviews.
'"I used to take another oral contraceptive, which had 21 pill cycle, and was very happy- very light periods, max 5 days, no other side effects. But it contained hormone gestodene, which is not available in US, so I switched to Lybrel, because the ingredients are similar. When my other pills ended, I started Lybrel immediately, on my first day of period, as the instructions said. And the period lasted for two weeks. When taking the second pack- same two weeks. And now, with third pack things got even worse- my third period lasted for two weeks and now it's the end of the third week- I still have daily brown discharge.\r\nThe positive side is that I didn't have any other side effects. The idea of being period free was so tempting... Alas."'
'"I have taken anti-depressants for years, with some improvement but mostly moderate to severe side affects, which makes me go off them.\r\n\r\nI only take Cymbalta now mostly for pain.\r\n\r\nWhen I began Deplin, I noticed a major improvement overnight. More energy, better disposition, and no sinking to the low lows of major depression. I have been taking it for about 3 months now and feel like a normal person for the first time ever. Best thing, no side effects."'
As we can observe from the sample reviews, it contains special symbols, “\r\n”, “”, etc.
So first we will remove double quotes from the dataset.
for i, col in enumerate(X.columns):
X.iloc[:, i] = X.iloc[:, i].str.replace('"', '')Next, we will create a review cleaning function that will remove HTML tags, lowercase the text reviews, retain only alphabets, remove stop words, and perform lemmatization.
from bs4 import BeautifulSoup
import re
from nltk.stem import WordNetLemmatizer
from nltk.corpus import stopwords
stop = stopwords.words('english')
lemmatizer = WordNetLemmatizer()
def review_to_words(raw_review):
# 1. Delete HTML
review_text = BeautifulSoup(raw_review, 'html.parser').get_text()
# 2. Make a space
letters_only = re.sub('[^a-zA-Z]', ' ', review_text)
# 3. lower letters
words = letters_only.lower().split()
# 5. Stopwords
meaningful_words = [w for w in words if not w in stop]
# 6. lemmitization
lemmitize_words = [lemmatizer.lemmatize(w) for w in meaningful_words]
# 7. space join words
return( ' '.join(lemmitize_words))
X['review_clean'] = X['review'].apply(review_to_words)
#creating feature and target variable
X_feat=X['review_clean']
y=X['condition']
X.head()
As we can see from the above output reviews are not cleaned properly which we can use for vectorizing using TF-IDF.
In the previous step, we have already created a feature and target variable. So, next, we will proceed to Train Test split.
X_train, X_test, y_train, y_test = train_test_split(X_feat, y,stratify=y,test_size=0.2, random_state=0)
So, we are using 80% of the data for training the model and rest 20% for testing the model.
Next, we will vectorize the data using TF-IDF and use Bi-grams for the same.
tfidf_vectorizer2 = TfidfVectorizer(max_df=0.8, ngram_range=(1,2)) tfidf_train_2 = tfidf_vectorizer2.fit_transform(X_train) tfidf_test_2 = tfidf_vectorizer2.transform(X_test)
In TfidfVectorizer we have used two parameters one is max_df =0.8 means the bigrams which come less than or equal to 80% of the time in the document will be considered for vectorization. The second one is ngram_range=(1,2) it means we want to consider both unigram and bi-grams for vectorizing the data.
Model Building
In the model building, we will use two main algorithms one is Multinomial Naive Bayes Algorithm as it usually works well on textual data and the other one is Passive-aggressive Classifier.
mnb_tf = MultinomialNB()
mnb_tf.fit(tfidf_train_2, y_train)
pred = mnb_tf.predict(tfidf_test_2)
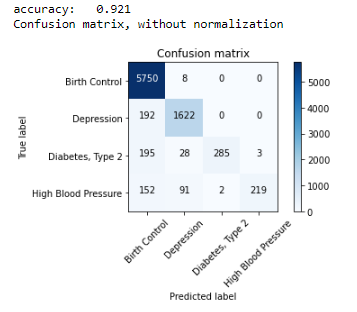
score = metrics.accuracy_score(y_test, pred)
print("accuracy: %0.3f" % score)
cm = metrics.confusion_matrix(y_test, pred, labels=['Birth Control', 'Depression','Diabetes, Type 2','High Blood Pressure'])
plot_confusion_matrix(cm, classes=['Birth Control', 'Depression','Diabetes, Type 2','High Blood Pressure'])
pass_tf = PassiveAggressiveClassifier()
pass_tf.fit(tfidf_train_2, y_train)
pred = pass_tf.predict(tfidf_test_2)
score = metrics.accuracy_score(y_test, pred)
print("accuracy: %0.3f" % score)
cm = metrics.confusion_matrix(y_test, pred, labels=['Birth Control', 'Depression','Diabetes, Type 2','High Blood Pressure'])
plot_confusion_matrix(cm, classes=['Birth Control', 'Depression','Diabetes, Type 2','High Blood Pressure'])
As we can see from the above results, Passive Aggressive Classifier accuracy is 6% more than Multinomial Naive Bayes Algorithm so we will be selecting it as our main model for medical condition prediction.
Most Informative Feature
Now, we will analyze the top 10 most informative features for a particular medical condition.
def most_informative_feature_for_class(vectorizer, classifier, classlabel, n=10):
labelid = list(classifier.classes_).index(classlabel)
feature_names = vectorizer.get_feature_names()
topn = sorted(zip(classifier.coef_[labelid], feature_names))[-n:]
for coef, feat in topn:
print (classlabel, feat, coef)
most_informative_feature_for_class(tfidf_vectorizer2, pass_tf, 'Birth Control')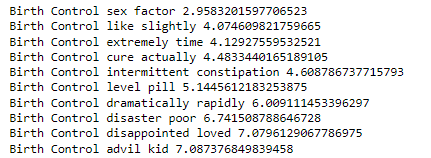
As per the output above, the bi-gram features seem relevant based on the medical condition “Birth Control”.
Similarly, we can analyze top informative features for other medical conditions.
Now, to predict medical condition and recommend drugs based on predicted condition and reviews rating and useful count, we will create two functions i.e., top_drugs_extractor and predict_text
## Function for Extracting Top drugs
def top_drugs_extractor(condition):
df_top = df[(df['rating']>=9)&(df['usefulCount']>=100)].sort_values(by = ['rating', 'usefulCount'], ascending = [False, False])
drug_lst = df_top[df_top['condition']==condition]['drugName'].head(3).tolist()
return drug_lst
def predict_text(lst_text):
df_test = pd.DataFrame(lst_text, columns = ['test_sent'])
df_test["test_sent"] = df_test["test_sent"].apply(review_to_words)
tfidf_bigram = tfidf_vectorizer3.transform(lst_text)
prediction = pass_tf.predict(tfidf_bigram)
df_test['prediction']=prediction
return df_test
# sample sentences for recommending drugs
sentences = [
"I have only been on Tekturna for 9 days. The effect was immediate. I am also on a calcium channel blocker (Tiazac) and hydrochlorothiazide. I was put on Tekturna because of palpitations experienced with Diovan (ugly drug in my opinion, same company produces both however). The palpitations were pretty bad on Diovan, 24 hour monitor by EKG etc. After a few days of substituting Tekturna for Diovan, there are no more palpitations.",
"This is the third med I've tried for anxiety and mild depression. Been on it for a week and I hate it so much. I am so dizzy, I have major diarrhea and feel worse than I started. Contacting my doc in the am and changing asap.",
"I just got diagnosed with type 2. My doctor prescribed Invokana and metformin from the beginning. My sugars went down to normal by the second week. I am losing so much weight. No side effects yet. Miracle medicine for me",]
tfidf_trigram = tfidf_vectorizer3.transform(sentences)
predictions = pass_tf.predict(tfidf_trigram)
for text, label in zip(sentences, predictions):
if label=="High Blood Pressure":
target="High Blood Pressure"
top_drugs = top_drugs_extractor(label)
print("text:", text, "\nCondition:", target)
print("Top 3 Suggested Drugs:")
print(top_drugs[0])
print(top_drugs[1])
print(top_drugs[2])
print()
elif label=="Depression":
target="Depression"
top_drugs = top_drugs_extractor(label)
print("text:", text, "\nCondition:", target)
print("Top 3 Suggested Drugs:")
print(top_drugs[0])
print(top_drugs[1])
print(top_drugs[2])
print()
elif label=="Diabetes, Type 2":
target="Diabetes, Type 2"
top_drugs = top_drugs_extractor(label)
print("text:", text, "\nCondition:", target)
print("Top 3 Suggested Drugs:")
print(top_drugs[0])
print(top_drugs[1])
print(top_drugs[2])
print()
else:
target="Birth Control"
print("text:", text, "\Condition:", target)
top_drugs = top_drugs_extractor(label)
print("text:", text, "\nCondition:", target)
print("Top 3 Suggested Drugs:")
print(top_drugs[0])
print(top_drugs[1])
print(top_drugs[2])
print()
As we can see from the above code we have successfully built a machine learning-based recommendation system for predicting medical conditions and recommending the top 3 relevant drugs based on predicted medical conditions and top reviews rating and useful reviews count.
I hope this article will help you in building end to end drug recommendations system. the whole code demonstrated in this article is available at this Git repo.
